# K-Means Clustering and k-means++ Initialization: A Practical Tutorial
🔍 Overview
K-Means is a popular clustering algorithm used to partition a dataset into K clusters by minimizing intra-cluster variance. A crucial factor in its performance is how you initialize the cluster centroids. This tutorial covers:
- The mechanics of K-Means and k-means++ initialization
- How to choose the right number of clusters
-
Best practices to avoid centroid collapse
🔧 K-Means Algorithm
Given a dataset $X = {x_1, x_2, \ldots, x_n} \subset \mathbb{R}^d$, K-Means partitions the data into $K$ clusters ${C_1, \ldots, C_K}$ by minimizing the total within-cluster sum of squares:
📌 Inertia (Objective Function)
The quantity minimized in k-means is called inertia:
\[\text{Inertia} = \sum_{i=1}^{K} \sum_{x_j \in C_i} \|x_j - c_i\|^2\]Where $c_i$ is the centroid of cluster $C_i$.
✅ k-means++ Initialization
📁 Problem with Random Initialization
Randomly selecting $K$ centroids can cause poor convergence, centroid collapse, or suboptimal clustering.
🔎 k-means++ Algorithm
A better strategy is k-means++, which chooses centroids to be diverse and far apart. Here’s the step-by-step:
- Choose the first centroid $c_1 \in X$ randomly.
-
For each remaining centroid $c_k$:
- For every $x \in X$, compute: $D(x)^2 = \min_{1 \leq j < k} |x - c_j|^2$
- Sample next centroid with probability: $P(x) = \frac{D(x)^2}{\sum_{x’ \in X} D(x’)^2}$
- Repeat until $K$ centroids are selected.
- Proceed with regular Lloyd’s algorithm.
⚡ Benefits
- Strong theoretical guarantees: expected cost $\leq 8(\log K + 2) \cdot \text{OPT}$
- Prevents collapse and encourages diverse cluster initialization
🧪 Python Code Example
Here’s a simple k-means implementation using NumPy with inline shape comments:
import numpy as np
def initialize_kmeans_plusplus(X, K):
# X: (n_samples, n_features)
n_samples = X.shape[0]
centroids = [X[np.random.randint(n_samples)]] # first centroid: (n_features,)
for _ in range(1, K):
# dists: (n_samples,), each element is D(x)^2
dists = np.array([min(np.linalg.norm(x - c)**2 for c in centroids) for x in X])
probs = dists / dists.sum() # (n_samples,)
cumulative_probs = np.cumsum(probs) # (n_samples,)
r = np.random.rand()
idx = np.searchsorted(cumulative_probs, r)
centroids.append(X[idx]) # (n_features,)
return np.array(centroids) # (K, n_features)
def kmeans(X, K, max_iters=100):
# X: (n_samples, n_features)
centroids = initialize_kmeans_plusplus(X, K) # (K, n_features)
for _ in range(max_iters):
# Compute pairwise distances: (n_samples, K)
# X[:, None]: (n_samples, 1, n_features)
# centroids[None, :]: (1, K, n_features)
labels = np.argmin(np.linalg.norm(X[:, None] - centroids[None, :], axis=2), axis=1) # (n_samples,)
# Update centroids: (K, n_features)
new_centroids = np.array([X[labels == k].mean(axis=0) for k in range(K)])
if np.allclose(centroids, new_centroids):
break
centroids = new_centroids
return centroids, labels # centroids: (K, n_features), labels: (n_samples,)
Since k-means++ does not choose $K$, you need to select it manually. Here are common strategies:
1. Elbow Method
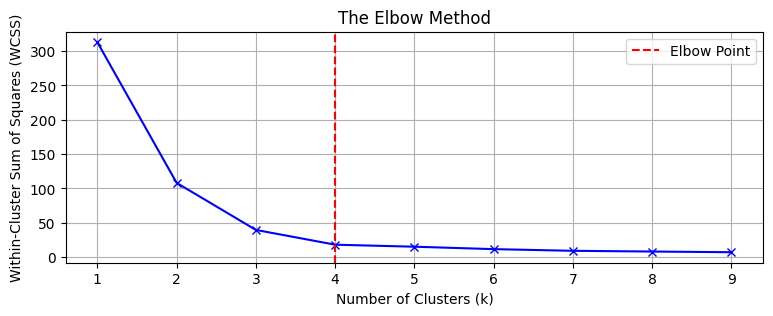
Fig. 1 Within-Cluster-Sum of Squares (WCSS) loss versus K
- Plot $K$ vs. inertia (WCSS)
- Look for an “elbow” where improvements slow down
2. Silhouette Score
- Measures separation vs. compactness:
- Choose $K$ that maximizes average silhouette
3. Gap Statistic
- Compares clustering performance to random uniform data
- Choose $K$ where gap is largest or flattens
4. Domain Knowledge
- Use task-specific or semantic insight to set $K$
📈 Summary Table
| Method | Output | Pros | Cons |
|---|---|---|---|
| Random Init | Centroids | Fast | High collapse risk |
| k-means++ | Centroids | Low collapse, better convergence | Slightly slower |
| Elbow | Optimal $K$ | Simple, visual | Subjective |
| Silhouette | Optimal $K$ | Captures quality of clustering | Expensive for large datasets |
| Gap Statistic | Optimal $K$ | Strong theoretical foundation | Computationally heavy |
| Domain Knowledge | Optimal $K$ | Expert-driven | May not always be available |
🚀 Next Steps
- Use k-means++ for better initialization
- Choose $K$ via elbow + silhouette for practical datasets
- Combine PCA/t-SNE visualization with clustering to validate

